Agenda: Hands-on Workshop on Computational Biophysics 2013
Agenda: Hands-on Workshop on Computational Biophysics
Workshop details: Homepage Instructors Evaluations
Mon, June 10: Introduction to Protein Structure and Dynamics - Klaus Schulten
| 08:00-08:30 | Registration and Continental Breakfast |
| 08:30-09:00 | Welcome and Brief Overview - Ivet Bahar (video) |
| 09:00-09:10 | Opening Remarks (video) |
| 09:10-10:30 | Structure and Sequence Analysis with VMD (video) |
| Coffee Break | |
| 10:50-12:00 | Introduction to Molecular Dynamics with NAMD (video) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 1400-16:00 | VMD Tutorial - Using VMD; NAMD Tutorial |
| Coffee Break | |
| `6:15-18:00 | VMD Tutorial - Using VMD; NAMD Tutorial |
Tue, June 11: Statistical Mechanics of Proteins; Force Field Parameterization- Klaus Schulten and Emad Tajkhorshid
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Applications of VMD / NAMD in Modern Research (video1, video2) |
| Coffee Break | |
| 10:50- noon | Topology, Parameters, and Structure Files (video) |
| 12:00-12:30 pm | Q & A; Group picture |
| Lunch Break | |
| 14:00-16:00 pm |
Participant tutorial options: |
| Coffee Break | |
| 16:15-18:00 pm | Participant tutorial options: 1) NAMD Tutorial & Stretching Deca-alanine, or 2) Expert NAMD Set Tutorials* 3) Parameterizing a Novel Residue 4) Topology File Tutorial |
| Dinner Break | (note: participants are on their own for this meal) |
| 19:00-21:00 | Presentations by workshop participants |
Wed, June 12: Simulating Membrane Proteins - Emad Tajkhorshid
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Introduction and Examples |
| Coffee Break | |
| 10:50-12:00 | Transport in Membrane Channels (video) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 14:00-16:30 | Participant tutorial options: 1) Membrane Proteins 2) Open tutorial time |
| Coffee Break | |
| 16:45-18:00 | Participant tutorial options: 1) Membrane Proteins 2) Open tutorial time |
Thu, June 13: Collective Dynamics of Proteins Using Elastic Network Models - Ivet Bahar, Tim Lezon and Ahmet Bakan
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Elastic Network Models (ENMs) and Collective Motions of Biomolecular Systems using ProDy (video1, video2) |
| Coffee Break | |
| 10:50-12:00 | ProDy Overview and Applications (PDF, video) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 14:00-16:00 | ProDy Tutorial; NMWiz Tutorial (PDF, video) |
| Coffee Break | |
| 16:15-18:00 |
Participant tutorial options: |
Fri, June 14: Druggability Simulations, and Analyzing Sequence Patterns and Structural Dynamics - Ivet Bahar and Ahmet Bakan
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Druggability: Method and Applications (PDF, video) |
| Coffee Break | |
| 10:50-12:00 | Comparative Analysis of Sequence Patterns and Structural Dynamics for Families/Ensembles of Proteins (PDF, video) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 13:30-15:00 | Druggability Tutorial; Evol Tutorial |
Tutorial Options:
* Expert NAMD Set Tutorials:
- Shape-Based Coarse Graining
- User-Defined Forces in NAMD
Click here to see descriptions of tutorials listed in program.
Note: participants may also download and get assistance with other tutorials from the TCBG tutorials website during the workshop.
Program subject to change. Workshop supported by NIH 9P41GM104601 "Center for Macromolecular Modeling and Bioinformatics", and NIH P41 GM103712 "NIH Center for Multiscale Modeling of Biological Systems".
Agenda: Hands-on Workshop on Computational Biophysics 2014
Workshop details: Homepage
Mon, May 19: Introduction to Protein Structure and Dynamics - Klaus Schulten
| 08:00-08:30 | Registration and Continental Breakfast |
| 08:30-09:00 | Welcome and Brief Overview - Ivet Bahar |
| 09:00-09:10 | Opening Remarks (video) |
| 09:10-10:30 | Structure and Sequence Analysis with VMD (PDF, video, video) |
| Coffee Break | |
| 10:50-12:00 | Introduction to Molecular Dynamics with NAMD (PDF, video) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 14:00-16:00 | VMD Tutorial - Using VMD; NAMD Tutorial |
| Coffee Break | |
| 16:15-18:00 | VMD Tutorial - Using VMD; NAMD Tutorial |
Tue, May 20: Statistical Mechanics of Proteins - Klaus Schulten
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Analysis of Equilibrium and Non-equilibrium Properties of Proteins with NAMD (PDF) |
| Coffee Break | |
| 10:50- 12:00 | Applications of VMD / NAMD in Modern Research (PDF, video) |
| 12:00-12:30 | Q & A; Group picture |
| Lunch Break | |
| 14:00-16:00 |
Participant tutorial options: |
| Coffee Break | |
| 16:15-18:00 | Participant tutorial options: 1) NAMD Tutorial & Stretching Deca-alanine, or 2) GPU Accelerated Molecular Dynamics and Visualization, or 3) Shape-Based Coarse Graining |
| Dinner Break |
(note: participants are on their own for this meal) |
| 19:00-21:00 | Presentations by workshop participants |
Wed, May 21: Computational Nano-Bio - Alek Aksimentiev
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Introduction to Modeling and Simulations of Nucleic Acid Systems (PDF, video) |
| Coffee Break | |
| 10:50-12:00 | Modeling the Interface Between Biological and Synthetic Materials (PDF, video) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 14:00-16:30 | Modeling Nanopores for Sequencing DNA; Introduction to MD Simulation of DNA-protein Systems |
| Coffee Break | |
| 16:45-18:00 | Modeling Nanopores for Sequencing DNA; Introduction to MD Simulation of DNA-protein Systems; |
Thu, May 22: Collective Dynamics of Proteins Using Elastic Network Models - Ivet Bahar, Tim Lezon and Chakra Chennubhotla
| 08:30-09:00 | Continental Breakfast |
| 09:00-10:30 | Elastic Network Models (ENMs) and Collective Motions of Biomolecular Systems using ProDy (video) |
| Coffee Break | |
| 10:50-12:00 | ProDy Overview and Applications (PDF) |
| 12:00-12:20 | Q & A |
| Lunch Break | |
| 14:00-16:00 | ProDy Tutorial; NMWiz Tutorial |
| Coffee Break | |
| 16:15-18:00 |
Participant tutorial options: |
Fri, May 23: Druggability Simulations and Sequence Evolution Patterns - Ivet Bahar, Indira Shrivastava, and Chakra Chennubhotla
| 08:30-09:00 | Continental Breakfast |
| 09:00-09:30 | Druggability: Method (PDF, video) |
| 09:30-10:30 | Druggability: Applications in ProDy |
| Coffee Break | |
| 10:50-11:20 | Comparative Analysis of Sequence Patterns and Structural Dynamics for Families/Ensembles of Proteins (PDF, video) |
| 11:20-12:30 | Comparative Sequence Analysis in ProDy |
| Lunch Break | |
| 13:30-15:00 | Druggability and Evol Tutorials in ProDy |
| Coffee Break | |
| 16:15-18:00 | Tutorial options |
Tutorials
Note: participants may also download and get assistance with other tutorials from the TCBG tutorials website, the Aksimentiev group tutorial pages and the ProDy tutorials website during the workshop.
Program subject to change. Workshop supported by NIH 9P41GM104601 "Center for Macromolecular Modeling and Bioinformatics" and NIH P41 GM103712 "NIH Center for Multiscale Modeling of Biological Systems".
Agenda: Hands-on Workshop on Computational Biophysics 2015
Agenda: Hands-on Workshop on Computational Biophysics 2015
Mon. June1: Introduction to Protein Structure and Dynamics - Klaus Schulten
| 08:00-08:30 | Registration and continental breakfast |
| 08:30-09:00 | Welcome and Brief Overview - Ivet Bahar |
| 09:00-09:10 | Opening Remarks (PDF, video) |
| 09:10-10:30 | Structure and Sequence Analysis with VMD (PDF, video) |
| Coffee break | |
| 10:50-12:00 | Introduction to Molecular Dynamics with NAMD (PDF, video) |
| 12:00-12:20 | Q & A |
| Lunch break | |
| 14:00-16:00 | VMD Tutorial - Using VMD; NAMD Tutorial |
| Coffee break | |
| 16:15-18:00 | VMD Tutorial - Using VMD; NAMD Tutorial |
Tue. June 2: Statistical Mechanics of Proteins - Klaus Schulten
| 08:30-09:00 | Continental breakfast | |
| 09:00-10:30 | Analysis of Equilibrium and Non-equilibrium Properties of Proteins with NAMD (PDF1, PDF2, video) | |
| Coffee break | ||
| 10:50- 12:00 | Applications of VMD / NAMD in Modern Research (PDF1, PDF2, video) | |
| 12:00-12:30 | Q & A; Group picture | |
| Lunch break | ||
| 14:00-16:00 |
Participant tutorial options: |
|
| Coffee break | ||
| 16:15-18:00 | Participant tutorial options: 1) NAMD Tutorial & Stretching Deca-alanine 2) GPU Accelerated Molecular Dynamics and Visualization 3) Shape-Based Coarse Graining |
|
| Dinner break (note: participants are on their own for this meal) |
||
| 19:00-21:00 | Presentations by workshop participants | |
Wed. June 3: Introductions to Evolutionary Concepts, Network Analysis,and Cell Simulations - Zan Luthey-Schulten
| 08:30-09:00 | Continental breakfast |
| 09:00-10:30 | Applications of Evolutionary Algorithms and Network Analysis in MultiSeq/VMD to Translation (PDF, video) |
| Coffee break | |
| 10:50-12:00 | Introduction to Simulations of Bacterial Cells (video) |
| 12:00-12:20 | Q & A |
| Lunch break | |
| 14:00-18:00 | Participant tutorial options: 1) Evolution of Translation 2) Network Analysis 3) Lattice Microbe Simulations |
Thu. June 4: Collective Dynamics of Proteins Using Elastic Network Models - Ivet Bahar, Tim Lezon and Chakra Chennubhotla
| 08:30-09:00 | Continental breakfast |
| 09:00-10:30 | Elastic Network Models (ENMs) and Collective Motions using ANM Server and ProDy API (PDF, video) |
| Coffee break | |
| 10:50-12:00 | ProDy Overview and Applications (PDF, video) |
| 12:00-12:20 | Q & A |
| Lunch break | |
| 14:00-16:00 | ProDy Tutorial; NMWiz Tutorial |
| Coffee break | |
| 16:15-18:00 |
Participant tutorial options: |
Fri. June 5: Druggability Simulations and Sequence Evolution Patterns - Ivet Bahar, Indira Shrivastava, and Cihan Kaya
| 08:30-09:00 | Continental breakfast |
| 09:00-10:30 | Druggability: Methods and Applications using ProDy (PDF, video) |
| Coffee break | |
| 10:50-12:00 | EVOL: Comparative Analysis of Sequence Evolution Patterns, Structure, and Dynamics (PDF, video) |
| 12:00-12:30 | Druggability tutorial using ProDy |
| Lunch break | |
| 14:00-15:30 | Evol Tutorials in ProDy |
| 15:30-16:00 | Discussion and Closure |
| Coffee break | |
| 16:30-18:00 | Additional tutorials (optional) |
Tutorials
Note: participants may also download and get assistance with other tutorials from the TCBG tutorials website and the ProDy tutorials website during the workshop.
Program subject to change. Workshop supported by NIH 9P41GM104601 "Center for Macromolecular Modeling and Bioinformatics" and NIH P41 GM103712 "NIH Center for Multiscale Modeling of Biological Systems".
Past Workshops
MMBios Partners present workshops throughout the year. Materials from past events are available shortly after workshops are completed unless otherwise noted.
-----
Hands-on Workshop on Computational Biophysics
Workshop dates: June 10-14, 2013
Location: Pittsburgh Supercomputing Center, 300 South Craig Street, Pittsburgh, PA 15213
This workshop will be presented by members of the BTRC Center for Multiscale Modeling of Biological Systems from the University of Pittsburgh and the members of the Theoretical and Computational Biophysics Group (www.ks.uiuc.edu) from the University of Illinois at Urbana-Champaign. It will cover a wide range of physical models and computational approaches for the simulation of biological systems including ProDy, NAMD and VMD. [Read More]
Computational Methods for Spatially Realistic Microphysiological Simulations
Workshop Dates: Mon, May 20 - Wed, May 22, 2013
Location: Pittsburgh Supercomputing Center, 300 South Craig Street, Pittsburgh, PA 15213
This workshop is organized by the Center for Multiscale Modeling of Biological Systems and will cover theory and practice for the design and simulation of models focused on diffusion-reaction systems such as neurotransmission, signaling cascades, and other forms of biochemical networks. The lastest version of the MCell simulation environment (www.mcell.org) will be introduced, including new Monte Carlo methods for 3-D simulation of reactions in solution and on arbitrarily shaped biological surfaces. [Read More]
Subcategories
Latest News
Upcoming Events
| No events |
Past Events
| No events |
Collaborate with MMBioS
Research Highlights
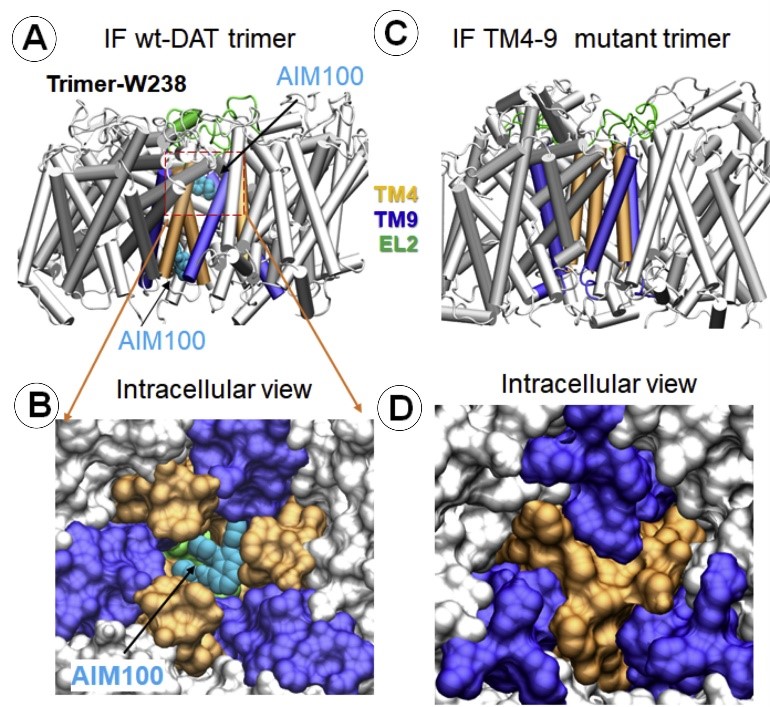
The Bahar (TR&D1) and Sorkin (DBP3) labs published an article in the Journal of Biological Chemistry, selected as one of JBC's "Editors' Picks. Our results demonstrate a direct coupling between conformational dynamics of DAT, functional activity of the transporter and its oligomerization leading to endocytosis. The high specificity of such coupling for DAT makes the TM4-9 hub a new target for pharmacological modulation of DAT activity and subcellular localization. (Read more)

Differences in the intrinsic spatial dynamics of the chromatin contribute to cell differentiation
Comparison with RNA-seq expression data reveals a strong overlap between highly expressed genes and those distinguished by high mobilities in the present study, in support of the role of the intrinsic spatial dynamics of chromatin as a determinant of cell differentiation. (Read more)

Nanoscale co-organization and coactivation of AMPAR, NMDAR, and mGluR at excitatory synapses
Work by TR&D2 Investigators and collaborators provide insights into the nanometer scale organization of postsynaptic glutamate receptors using a combination of dual-color superresolution imaging, electrophysiology, and computational modeling. (Read more)

Parallel Tempering with Lasso for model reduction in systems biology
TR&D3 Investigators and collaborators develop PTLasso, a Bayesian model reduction approach that combines Parallel Tempering with Lasso regularization, to automatically extract minimal subsets of detailed models that are sufficient to explain experimental data. On both synthetic and real biological data, PTLasso is an effective method to isolate distinct parts of a larger signaling model that are sufficient for specific data. (Read more)

Image-derived models of cell organization changes during differentiation and drug treatments
Our work on modeling PC12 cells undergoing differentiation into neuron-like morphologies (under C&SP11, completed) has been published in Molecular Biology of the Cell. We have also made the large dataset of 3D images collected in that study available through Dryad. (Read more)

Monoamine transporters: structure, intrinsic dynamics and allosteric regulation
T&RD1 investigators Mary Cheng and Ivet Bahar published an invited review article in Nature Structural & Molecular Biology, addressing recent progress in the elucidation of the structural dynamics of MATs and their conformational landscape and transitions, as well as allosteric regulation mechanisms. (Read more)

Trimerization of dopamine transporter triggered by AIM-100 binding
The Bahar (TR&D1) and Sorkin (DBP3) labs explored the trimerization of dopamine transporter (DAT) triggered by a furopyrimidine, AIM-100, using a combination of computational and biochemical methods, and single-molecule live-cell imaging assays. (Read more)

Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency
TR&D2 investigators and collaborators describe organizing role of neuroligin-1 to align post-synaptic AMPA Receptors with pre-synaptic release sites into trans-synaptic “nano-columns” to enhance signaling.(Read more)

Inferring the Assembly Network of Influenza Virus
In an article in PLoS Computational Biology, MMBioS TR&D4 members Xiongto Ruan and Bob Murphy collaborated with Seema Lakdawala to address this question of the assembly network of the Influenza virus.(Read more)

Our findings highlight an important mechanism by which proteins genetically implicated in Parkinson’s disease (PD; PINK1) and frontotemporal dementia (FTD; VCP) interact to support the health and maintenance of neuronal arbors.(Read more)

Improved methods for modeling cell shape
In a recent paper in Bioinformatics, Xiongtao Ruan and Bob Murphy of TR&D4 addressed the question of how best to model cell and nuclear shape.(Read more)

New tool to predict pathogenicity of missense variants based on structural dynamics: RHAPSODY
We demonstrated that the analysis of a protein’s intrinsic dynamics can be successfully used to improve the prediction of the effect of point mutations on a protein functionality. This method employs ANM/GNM tools (Read more)

New method for investigating chromatin structural dynamics.
By adapting the Gaussian Network Model (GNM) protein-modeling framework, we were able to model chromatin dynamics using Hi-C data, which led to the identification of novel cross-correlated distal domains (CCDDs) that were found to also be associated with increased gene co-expression. (Read more)

Structural elements coupling anion conductance and substrate transport identified
We identified an intermediate anion channeling state (iChS) during the global transition from the outward facing (OF) to inward facing state (IFS). Our prediction was tested and validated by experimental study conducted in the Amara lab (NIMH). Critical residues and interactions were analyzed by SCAM, electrophysiology and substrate uptake experiments (Read more)

Integrating MMBioS technologies for multiscale discovery
TR&D teams driven by individual DBPs are naturally joining forces, integrating their tools to respond to the needs of the DBP, and creating integrative frameworks for combining structural and kinetic data and computing technologies at multiple scales. (Read more)

Large scale visualization of rule-based models.
Signaling in living cells is mediated through a complex network of chemical interactions. Current predictive models of signal pathways have hundreds of reaction rules that specify chemical interactions, and a comprehensive model of a stem cell or cancer cell would be expected to have many more. Visualizations of rules and their interactions are needed to navigate, organize, communicate and analyze large signaling models. (Read more)

Integration of MCellR into MCell/CellBlender
Using spatial biochemical models of SynGAP/PSD95, MMBioS investigators were able to merge the MCellR code-base with the MCell code-base and validate its utility and correctness of this sophisticated technology now easily accessible through the MCell/CellBlender GUI. (Read more)
Causal relationships of spatial distributions of T cell signaling proteins
The idea is to identify a relationship in which a change in the concentration of one protein in one cell region consistently is associated with a change in the concentration of another protein in the same or a different region. We used the data from our Science Signaling paper reported last year to construct a model for T cells undergoing stimulation by both the T cell receptor and the costimulatory receptor. (Read more...)

BioNetGen modeling helps reveal immune system response decision
To attack or to let be is an important decision that our immune systems must make to protect our bodies from foreign invaders or protect bodily tissues from an immune attack. Using modeling and experiments, we have painted a sharper picture of how T cells make these critical decisions. (Read more)

Tools for determining the spatial relationships between different cell components
An important task for understanding how cells are organized is determining which components have spatial patterns that are related to each other.Read more

Pipeline for creation of spatiotemporal maps
Using a combination of diffeomorphic methods and improved cell segmentation, we developed a CellOrganizer pipeline for use in DPB4 to construct models of the 4D distributions of actin and 8 of its regulators during the response of T cells to antigen presentation. Read more

Multi-scale Hybrid Methodology
The hybrid methodology, coMD, that we have recently developed [1] has been recently extended to construct the energy landscape near the functional states of LeuT (Fig 1) [2]. This is the first energy landscape constructed for this NSS family member. Read more

Insights into the cooperative dynamics of AMPAR
Comparative analysis of AMPAR and NMDAR dynamics reveals striking similarities, opening the way to designing new modulators of allosteric interactions. Read more

Improved Sampling of Cell-Scale Models using the WE Strategy
The WE strategy for orchestrating a large set of parallel simulations has now been extended to spatially resolved cell-scale systems. The WESTPA implementation of WE has been used to control MCell simulations, including models built using a BioNetGen-CellOrganizer pipeline for situating complex biochemistry within spatially realistic cell models. Read more

Anatomy and Function of an Excitatory Network in the Visual Cortex
MMBioS researcher Greg Hood’s collaboration with Wei-Chung Allen Lee of Harvard University and R. Clay Reid of the Allen Institute for Brain Science concerning the reconstruction of an excitatory nerve-cell network in the mouse brain cortex at a subcellular level using the AlignTK software has been published in Nature. Read more
 Molecular Mechanism of Dopamine Transport by hDAT
Molecular Mechanism of Dopamine Transport by hDAT
Dopamine transporters (DATs) control neurotransmitter dopamine (DA) homeostasis by reuptake of excess DA, assisted by sodium and chloride ions. The recent resolution of DAT structure (dDAT) from Drosophila permits us for the first time to directly view the sequence of events involved in DA reuptake in human DAT (hDAT). Read more
 Synaptic Facilitation Revealed
Synaptic Facilitation Revealed
An investigation of several mechanisms of short-term facilitation at the frog neuromuscular junction concludes that the presence of a second class of calcium sensor proteins distinct from synaptotagmin can explain known properties of facilitation. Read more
 Sparse Graphical Models of Protein:Protein Interactions
Sparse Graphical Models of Protein:Protein Interactions
DgSpi is a new method for learning and using graphical models that explicitly represent the amino acid basis for interaction specificity and extend earlier classification-oriented approaches to predict ΔG of binding. Read more
 Advancing Parallel Bio-simulations
Advancing Parallel Bio-simulations
A new non-Markovian analysis can eliminate bias in estimates of long-timescale behavior, such as the mean first-passage time for the dissociation of methane molecules in explicit solvent. Read more